#include <BALL/QSAR/kernel.h>
Public Member Functions | |
Constructors and Destructors | |
| Kernel (Model *m, int k_type, double p1, double p2=-1) | |
| Kernel (Model *m, String f, String g) | |
| Kernel (Model *m, Vector< double > &w) | |
| Kernel (Model *m, const LinearModel &lm, int column) | |
| ~Kernel () | |
Friends | |
| class | FeatureSelection |
Attributes | |
|
| |
| int | type |
| double | par1 |
| double | par2 |
| String | equation1 |
| String | equation2 |
| Model * | model_ |
| Vector< double > | weights_ |
Accessors | |
|
| |
| void | calculateKernelMatrix (Matrix< double > &input, Matrix< double > &output) |
| void | calculateKernelMatrix (Matrix< double > &K, Matrix< double > &m1, Matrix< double > &m2, Matrix< double > &output) |
| void | calculateKernelVector (Matrix< double > &K, Vector< double > &m1, Matrix< double > &m2, Vector< double > &output) |
| void | gridSearch (double step_width, int steps, int recursions, int k, bool opt=0) |
| void | gridSearch (double step_width, int steps, bool first_rec, int k, double par1_start, double par2_start, bool opt) |
| void | calculateWeightedKernelMatrix (Matrix< double > &input, Matrix< double > &output) |
| void | calculateWeightedKernelMatrix (Matrix< double > &m1, Matrix< double > &m2, Matrix< double > &output) |
| void | calculateKernelMatrix1 (Matrix< double > &input, Matrix< double > &output) |
| void | calculateKernelMatrix2 (Matrix< double > &input, Matrix< double > &output) |
| void | calculateKernelMatrix3 (Matrix< double > &input, Matrix< double > &output) |
| void | calculateKernelMatrix4 (Matrix< double > &input, Matrix< double > &output) |
| void | calculateKernelMatrix1 (Matrix< double > &m1, Matrix< double > &m2, Matrix< double > &output) |
| void | calculateKernelMatrix2 (Matrix< double > &m1, Matrix< double > &m2, Matrix< double > &output) |
| void | calculateKernelMatrix3 (Matrix< double > &m1, Matrix< double > &m2, Matrix< double > &output) |
| void | calculateKernelMatrix4 (Matrix< double > &m1, Matrix< double > &m2, Matrix< double > &output) |
Definition at line 53 of file QSAR/kernel.h.
| BALL::QSAR::Kernel::Kernel | ( | Model * | m, | |
| const LinearModel & | lm, | |||
| int | column | |||
| ) |
constructor for weighted distance kernel.
| column | no of column of LinearModel.training_result that is to be used as weights vector |
| BALL::QSAR::Kernel::~Kernel | ( | ) |
| void BALL::QSAR::Kernel::calculateKernelMatrix | ( | Matrix< double > & | K, | |
| Matrix< double > & | m1, | |||
| Matrix< double > & | m2, | |||
| Matrix< double > & | output | |||
| ) |
calculates pairwise distance between all substances of m1 and m2 and saves them to Matrix<double> output.
If Kernel.weights is not empty, function Kernel.calculateWeightedDistanceMatrix() is used
Esle if: Kernel.f=="" and Kernel.g="", the distance between two substances a and b is calculated as 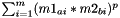 , with m=#descriptors
, with m=#descriptors
Else: distance is calculated as 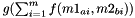
| void BALL::QSAR::Kernel::calculateKernelMatrix | ( | Matrix< double > & | input, | |
| Matrix< double > & | output | |||
| ) |
calculates pairwise distances between all substances in Matrix<double> input and saves them to Matrix<double> output.
If Kernel.weights is not empty, function Kernel.calculateWeightedDistanceMatrix() is used
Else if: Kernel.f=="" and Kernel.g="", the distance between two substances a and b is calculated as 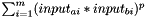 , with m=#descriptors
, with m=#descriptors
Else: distance is calculated as 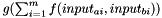
| void BALL::QSAR::Kernel::calculateKernelMatrix1 | ( | Matrix< double > & | m1, | |
| Matrix< double > & | m2, | |||
| Matrix< double > & | output | |||
| ) | [protected] |
| void BALL::QSAR::Kernel::calculateKernelMatrix1 | ( | Matrix< double > & | input, | |
| Matrix< double > & | output | |||
| ) | [protected] |
| void BALL::QSAR::Kernel::calculateKernelMatrix2 | ( | Matrix< double > & | m1, | |
| Matrix< double > & | m2, | |||
| Matrix< double > & | output | |||
| ) | [protected] |
| void BALL::QSAR::Kernel::calculateKernelMatrix2 | ( | Matrix< double > & | input, | |
| Matrix< double > & | output | |||
| ) | [protected] |
| void BALL::QSAR::Kernel::calculateKernelMatrix3 | ( | Matrix< double > & | m1, | |
| Matrix< double > & | m2, | |||
| Matrix< double > & | output | |||
| ) | [protected] |
| void BALL::QSAR::Kernel::calculateKernelMatrix3 | ( | Matrix< double > & | input, | |
| Matrix< double > & | output | |||
| ) | [protected] |
| void BALL::QSAR::Kernel::calculateKernelMatrix4 | ( | Matrix< double > & | m1, | |
| Matrix< double > & | m2, | |||
| Matrix< double > & | output | |||
| ) | [protected] |
| void BALL::QSAR::Kernel::calculateKernelMatrix4 | ( | Matrix< double > & | input, | |
| Matrix< double > & | output | |||
| ) | [protected] |
| void BALL::QSAR::Kernel::calculateKernelVector | ( | Matrix< double > & | K, | |
| Vector< double > & | m1, | |||
| Matrix< double > & | m2, | |||
| Vector< double > & | output | |||
| ) |
transforms test data 'input' into the kernel-saves and saves it to matrix 'output'
| void BALL::QSAR::Kernel::calculateWeightedKernelMatrix | ( | Matrix< double > & | m1, | |
| Matrix< double > & | m2, | |||
| Matrix< double > & | output | |||
| ) | [protected] |
calculates pairwise distances between all substances of m1 and m2, weighted by the contribution of every descriptor (as encoded in Kernel.weights), and saves them to Matrix<double> output.
Distance between two substances a and b is calculated as 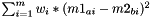 , with m=#descriptors
, with m=#descriptors
| void BALL::QSAR::Kernel::calculateWeightedKernelMatrix | ( | Matrix< double > & | input, | |
| Matrix< double > & | output | |||
| ) | [protected] |
calculates pairwise distances between all substances in Matrix<double> input, weighted by the contribution of every descriptor (as encoded in Kernel.weights), and saves them to Matrix<double> output.
Distance between two substances a and b is calculated as 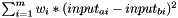 , with m=#descriptors
, with m=#descriptors
| void BALL::QSAR::Kernel::gridSearch | ( | double | step_width, | |
| int | steps, | |||
| bool | first_rec, | |||
| int | k, | |||
| double | par1_start, | |||
| double | par2_start, | |||
| bool | opt | |||
| ) | [protected] |
| void BALL::QSAR::Kernel::gridSearch | ( | double | step_width, | |
| int | steps, | |||
| int | recursions, | |||
| int | k, | |||
| bool | opt = 0 | |||
| ) |
grid search for the best kernel parameters.
Grid search is done locally around the current kernel parameter value(s).
| opt | if ==1, Model.optitimizeParameters() is used in each step of grid search, optimizing the parameter of the *Model* in addition to those of the kernel. | |
| step_width | the size of each step to be made | |
| steps | the number of steps for grid search | |
| recursions | number of recursions of grid search; in each recursion the step width is decreased by factor of 10 and searching is done in 20 steps around the values of the best kernel parameters determined in last recursion |
friend class FeatureSelection [friend] |
Definition at line 176 of file QSAR/kernel.h.
Equation for distance function for calculation of kernel matrix.
Distance of two substances a and b is calculated as 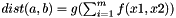 , with m=#descriptors
, with m=#descriptors
Use "x1" and "x2" in the String, e.g. "x1*x2"
Definition at line 121 of file QSAR/kernel.h.
Equation for distance function for calculation of kernel matrix.
Distance of two substances a and b is calculated as 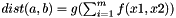 , with m=#descriptors
, with m=#descriptors
g determines what is to be done with the calculated "sum" over all elements (use "sum" in String); e.g. "sum^0.5" => euclidean distance if f=="x1*x2"
Definition at line 126 of file QSAR/kernel.h.
Model* BALL::QSAR::Kernel::model_ [protected] |
pointer to the model which uses this kernel
Definition at line 170 of file QSAR/kernel.h.
parameters for kernel functions set by the user
Definition at line 116 of file QSAR/kernel.h.
Definition at line 116 of file QSAR/kernel.h.
specifies which kind of kernel is chosen:
1 = polynomial kernel
2 = radial basis function kernel
3 = sigmoid kernel
4 = individual kernel-function
5 = weighted distance kernel
Definition at line 113 of file QSAR/kernel.h.
Vector<double> BALL::QSAR::Kernel::weights_ [protected] |
Definition at line 172 of file QSAR/kernel.h.
 1.6.3
1.6.3