#include <BALL/QSAR/gpModel.h>
Public Member Functions | |
Constructors and Destructors | |
| GPModel (const QSARData &q, int k_type, double p1, double p2=-1) | |
| GPModel (const QSARData &q, Vector< double > &w) | |
| GPModel (const QSARData &q, String s1, String s2) | |
| GPModel (const QSARData &q, const LinearModel &lm, int column) | |
| ~GPModel () | |
Accessors | |
| void | train () |
| Vector< double > | predict (const vector< double > &substance, bool transform=1) |
| double | calculateStdErr () |
| void | setParameters (vector< double > &v) |
| vector< double > | getParameters () const |
Private Attributes | |
Attributes | |
| Matrix< double > | L_ |
| Vector< double > | input_ |
| Vector< double > | K_t_ |
| double | lambda_ |
class for gaussian process regression
Definition at line 41 of file gpModel.h.
constructor that sets KernelModel.f to s1 and KernelModel.g to s2
| BALL::QSAR::GPModel::GPModel | ( | const QSARData & | q, | |
| const LinearModel & | lm, | |||
| int | column | |||
| ) |
| BALL::QSAR::GPModel::~GPModel | ( | ) |
| double BALL::QSAR::GPModel::calculateStdErr | ( | ) | [virtual] |
calculates standart error for the last prediction as 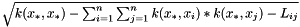
Reimplemented from BALL::QSAR::Model.
| vector<double> BALL::QSAR::GPModel::getParameters | ( | ) | const [virtual] |
Reimplemented from BALL::QSAR::Model.
| Vector<double> BALL::QSAR::GPModel::predict | ( | const vector< double > & | substance, | |
| bool | transform = 1 | |||
| ) | [virtual] |
Predicts the activities of a given substance
| substance | the substance which activity is to be predicted in form of a vecor containing the values for *all* descriptors (if neccessary, relevant descriptors will be selected automatically) | |
| transform | determines whether the values for each descriptor of the given substance should be transformed before prediction of activity. If (transform==1): each descriptor value is transformed according to the centering of the respective column of QSARData.descriptor_matrix used to train this model. If the substance to be predicted is part of the same input data (e.g. same SD-file) as the training data (as is the case during cross validation), transform should therefore be set to 0. |
Reimplemented from BALL::QSAR::KernelModel.
| void BALL::QSAR::GPModel::setParameters | ( | vector< double > & | ) | [virtual] |
sets the model parameters according to the given values.
Reimplemented from BALL::QSAR::Model.
| void BALL::QSAR::GPModel::train | ( | ) | [virtual] |
Starts training the model.
Implements BALL::QSAR::Model.
Vector<double> BALL::QSAR::GPModel::input_ [private] |
Vector<double> BALL::QSAR::GPModel::K_t_ [private] |
Matrix<double> BALL::QSAR::GPModel::L_ [private] |
double BALL::QSAR::GPModel::lambda_ [private] |
 1.6.3
1.6.3