BALL::QSAR::Kernel Class Reference
#include <BALL/QSAR/kernel.h>
Public Member Functions | |
Constructors and Destructors | |
| Kernel (Model *m, int k_type, double p1, double p2=-1) | |
| Kernel (Model *m, String f, String g) | |
| Kernel (Model *m, Vector< double > &w) | |
| Kernel (Model *m, const LinearModel &lm, int column) | |
| ~Kernel () | |
Friends | |
| class | FeatureSelection |
Attributes | |
|
| |
| int | type |
| double | par1 |
| double | par2 |
| String | equation1 |
| String | equation2 |
| Model * | model_ |
| Vector< double > | weights_ |
Accessors | |
|
| |
| void | calculateKernelMatrix (Matrix< double > &input, Matrix< double > &output) |
| void | calculateKernelMatrix (Matrix< double > &K, Matrix< double > &m1, Matrix< double > &m2, Matrix< double > &output) |
| void | calculateKernelVector (Matrix< double > &K, Vector< double > &m1, Matrix< double > &m2, Vector< double > &output) |
| void | gridSearch (double step_width, int steps, int recursions, int k, bool opt=0) |
| void | gridSearch (double step_width, int steps, bool first_rec, int k, double par1_start, double par2_start, bool opt) |
| void | calculateWeightedKernelMatrix (Matrix< double > &input, Matrix< double > &output) |
| void | calculateWeightedKernelMatrix (Matrix< double > &m1, Matrix< double > &m2, Matrix< double > &output) |
| void | calculateKernelMatrix1 (Matrix< double > &input, Matrix< double > &output) |
| void | calculateKernelMatrix2 (Matrix< double > &input, Matrix< double > &output) |
| void | calculateKernelMatrix3 (Matrix< double > &input, Matrix< double > &output) |
| void | calculateKernelMatrix4 (Matrix< double > &input, Matrix< double > &output) |
| void | calculateKernelMatrix1 (Matrix< double > &m1, Matrix< double > &m2, Matrix< double > &output) |
| void | calculateKernelMatrix2 (Matrix< double > &m1, Matrix< double > &m2, Matrix< double > &output) |
| void | calculateKernelMatrix3 (Matrix< double > &m1, Matrix< double > &m2, Matrix< double > &output) |
| void | calculateKernelMatrix4 (Matrix< double > &m1, Matrix< double > &m2, Matrix< double > &output) |
Detailed Description
Definition at line 53 of file QSAR/kernel.h.
Constructor & Destructor Documentation
| BALL::QSAR::Kernel::Kernel | ( | Model * | m, | |
| const LinearModel & | lm, | |||
| int | column | |||
| ) |
constructor for weighted distance kernel.
- Parameters:
-
column no of column of LinearModel.training_result that is to be used as weights vector
| BALL::QSAR::Kernel::~Kernel | ( | ) |
Member Function Documentation
| void BALL::QSAR::Kernel::calculateKernelMatrix | ( | Matrix< double > & | K, | |
| Matrix< double > & | m1, | |||
| Matrix< double > & | m2, | |||
| Matrix< double > & | output | |||
| ) |
calculates pairwise distance between all substances of m1 and m2 and saves them to Matrix<double> output.
If Kernel.weights is not empty, function Kernel.calculateWeightedDistanceMatrix() is used
Esle if: Kernel.f=="" and Kernel.g="", the distance between two substances a and b is calculated as 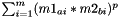 , with m=#descriptors
, with m=#descriptors
Else: distance is calculated as 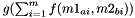
| void BALL::QSAR::Kernel::calculateKernelMatrix | ( | Matrix< double > & | input, | |
| Matrix< double > & | output | |||
| ) |
calculates pairwise distances between all substances in Matrix<double> input and saves them to Matrix<double> output.
If Kernel.weights is not empty, function Kernel.calculateWeightedDistanceMatrix() is used
Else if: Kernel.f=="" and Kernel.g="", the distance between two substances a and b is calculated as 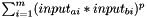 , with m=#descriptors
, with m=#descriptors
Else: distance is calculated as 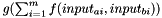
| void BALL::QSAR::Kernel::calculateKernelMatrix1 | ( | Matrix< double > & | m1, | |
| Matrix< double > & | m2, | |||
| Matrix< double > & | output | |||
| ) | [protected] |
| void BALL::QSAR::Kernel::calculateKernelMatrix1 | ( | Matrix< double > & | input, | |
| Matrix< double > & | output | |||
| ) | [protected] |
| void BALL::QSAR::Kernel::calculateKernelMatrix2 | ( | Matrix< double > & | m1, | |
| Matrix< double > & | m2, | |||
| Matrix< double > & | output | |||
| ) | [protected] |
| void BALL::QSAR::Kernel::calculateKernelMatrix2 | ( | Matrix< double > & | input, | |
| Matrix< double > & | output | |||
| ) | [protected] |
| void BALL::QSAR::Kernel::calculateKernelMatrix3 | ( | Matrix< double > & | m1, | |
| Matrix< double > & | m2, | |||
| Matrix< double > & | output | |||
| ) | [protected] |
| void BALL::QSAR::Kernel::calculateKernelMatrix3 | ( | Matrix< double > & | input, | |
| Matrix< double > & | output | |||
| ) | [protected] |
| void BALL::QSAR::Kernel::calculateKernelMatrix4 | ( | Matrix< double > & | m1, | |
| Matrix< double > & | m2, | |||
| Matrix< double > & | output | |||
| ) | [protected] |
| void BALL::QSAR::Kernel::calculateKernelMatrix4 | ( | Matrix< double > & | input, | |
| Matrix< double > & | output | |||
| ) | [protected] |
| void BALL::QSAR::Kernel::calculateKernelVector | ( | Matrix< double > & | K, | |
| Vector< double > & | m1, | |||
| Matrix< double > & | m2, | |||
| Vector< double > & | output | |||
| ) |
transforms test data 'input' into the kernel-saves and saves it to matrix 'output'
| void BALL::QSAR::Kernel::calculateWeightedKernelMatrix | ( | Matrix< double > & | m1, | |
| Matrix< double > & | m2, | |||
| Matrix< double > & | output | |||
| ) | [protected] |
calculates pairwise distances between all substances of m1 and m2, weighted by the contribution of every descriptor (as encoded in Kernel.weights), and saves them to Matrix<double> output.
Distance between two substances a and b is calculated as 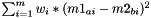 , with m=#descriptors
, with m=#descriptors
| void BALL::QSAR::Kernel::calculateWeightedKernelMatrix | ( | Matrix< double > & | input, | |
| Matrix< double > & | output | |||
| ) | [protected] |
calculates pairwise distances between all substances in Matrix<double> input, weighted by the contribution of every descriptor (as encoded in Kernel.weights), and saves them to Matrix<double> output.
Distance between two substances a and b is calculated as 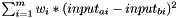 , with m=#descriptors
, with m=#descriptors
| void BALL::QSAR::Kernel::gridSearch | ( | double | step_width, | |
| int | steps, | |||
| bool | first_rec, | |||
| int | k, | |||
| double | par1_start, | |||
| double | par2_start, | |||
| bool | opt | |||
| ) | [protected] |
| void BALL::QSAR::Kernel::gridSearch | ( | double | step_width, | |
| int | steps, | |||
| int | recursions, | |||
| int | k, | |||
| bool | opt = 0 | |||
| ) |
grid search for the best kernel parameters.
Grid search is done locally around the current kernel parameter value(s).
- Parameters:
-
opt if ==1, Model.optitimizeParameters() is used in each step of grid search, optimizing the parameter of the *Model* in addition to those of the kernel. step_width the size of each step to be made steps the number of steps for grid search recursions number of recursions of grid search; in each recursion the step width is decreased by factor of 10 and searching is done in 20 steps around the values of the best kernel parameters determined in last recursion
Friends And Related Function Documentation
friend class FeatureSelection [friend] |
Definition at line 176 of file QSAR/kernel.h.
Member Data Documentation
Equation for distance function for calculation of kernel matrix.
Distance of two substances a and b is calculated as 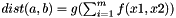 , with m=#descriptors
, with m=#descriptors
Use "x1" and "x2" in the String, e.g. "x1*x2"
Definition at line 121 of file QSAR/kernel.h.
Equation for distance function for calculation of kernel matrix.
Distance of two substances a and b is calculated as 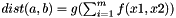 , with m=#descriptors
, with m=#descriptors
g determines what is to be done with the calculated "sum" over all elements (use "sum" in String); e.g. "sum^0.5" => euclidean distance if f=="x1*x2"
Definition at line 126 of file QSAR/kernel.h.
Model* BALL::QSAR::Kernel::model_ [protected] |
pointer to the model which uses this kernel
Definition at line 170 of file QSAR/kernel.h.
parameters for kernel functions set by the user
Definition at line 116 of file QSAR/kernel.h.
Definition at line 116 of file QSAR/kernel.h.
specifies which kind of kernel is chosen:
1 = polynomial kernel
2 = radial basis function kernel
3 = sigmoid kernel
4 = individual kernel-function
5 = weighted distance kernel
Definition at line 113 of file QSAR/kernel.h.
Vector<double> BALL::QSAR::Kernel::weights_ [protected] |
Definition at line 172 of file QSAR/kernel.h.
 1.6.3
1.6.3