NMR spectra and related classes
[Nuclear Magnetic
Resonance Spectroscopy]
Collaboration diagram for NMR spectra and related classes:
 |
Classes |
|
| class | BALL::AssignShiftProcessor |
| Set a property called
chemical_shift. More... |
|
| class | BALL::CreateSpectrumProcessor |
| Processor
creates peaklist_. More... |
|
| class | BALL::Experiment< PeakListType > |
| Class describing a certain type of
NMR experiment. More... |
|
| class | BALL::SimpleExperiment1D |
| Simple 1D NMR experiment class.
More... |
|
| class | BALL::Peak< PositionType > |
| Generic Peak Class. More... |
|
| class | BALL::PeakList< PT > |
| A generic peak list for spectra of
arbitrary dimension. More... |
|
| class | BALL::Spectrum< DataT, PeakT, PositionT > |
| A generic NMR spectrum class.
More... |
|
Functions |
|
| BALL_EXPORT const RegularData1D & | BALL::operator<< (RegularData1D &data, const PeakList1D &peak_list) |
| Create a simulated spectrum from a
peak list. |
|
| typedef Spectrum < RegularData1D, Peak1D > |
BALL::Spectrum1D |
| Convenience typedefs. |
|
|
typedef Spectrum < RegularData1D, Peak2D > |
BALL::Spectrum2D |
| Two-dimensional spectrum. |
|
|
typedef Spectrum < RegularData1D, Peak3D > |
BALL::Spectrum3D |
| Three-dimensional
spectrum. |
|
Typedef Documentation
| typedef Spectrum<RegularData1D, Peak1D> BALL::Spectrum1D |
Convenience typedefs.
One-dimensional spectrum
Function Documentation
| BALL_EXPORT const RegularData1D& BALL::operator<< | ( | RegularData1D & | data, | |
| const PeakList1D & | peak_list | |||
| ) |
Create a simulated spectrum from a peak list.
Using this operator, a peak list is converted to a gridded representation of the spectrum. The spectrum synthesis is based upon the assumption of a Lorentzian line shape. Peak width, position, and height are taken from each individual peak of the peak list. Each point in the RegularData1D array is assigned the sum of all Lorentzians centered at the peak positions:
![\[ S(\delta) = \sum_{i} \frac{h_i}{w_i (\delta_i-\delta)^2} \]](form_69.png)
where 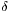 is the shift coordinate and each peak is
defined by its position
is the shift coordinate and each peak is
defined by its position 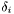 , intensity
, intensity 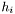 , and width
, and width
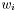 .
.
 1.5.8
1.5.8