 |
OpenMS
|
 |
OpenMS
|
#include <OpenMS/FEATUREFINDER/FeatureFinderMultiplexAlgorithm.h>
Public Member Functions | |
| FeatureFinderMultiplexAlgorithm () | |
| default constructor | |
| void | run (MSExperiment &exp, bool progress) |
| main method for feature detection | |
| FeatureMap & | getFeatureMap () |
| get methods | |
| ConsensusMap & | getConsensusMap () |
| MSExperiment & | getBlacklist () |
 Public Member Functions inherited from DefaultParamHandler Public Member Functions inherited from DefaultParamHandler | |
| DefaultParamHandler (const String &name) | |
| Constructor with name that is displayed in error messages. | |
| DefaultParamHandler (const DefaultParamHandler &rhs) | |
| Copy constructor. | |
| virtual | ~DefaultParamHandler () |
| Destructor. | |
| DefaultParamHandler & | operator= (const DefaultParamHandler &rhs) |
| Assignment operator. | |
| virtual bool | operator== (const DefaultParamHandler &rhs) const |
| Equality operator. | |
| void | setParameters (const Param ¶m) |
| Sets the parameters. | |
| const Param & | getParameters () const |
| Non-mutable access to the parameters. | |
| const Param & | getDefaults () const |
| Non-mutable access to the default parameters. | |
| const String & | getName () const |
| Non-mutable access to the name. | |
| void | setName (const String &name) |
| Mutable access to the name. | |
| const std::vector< String > & | getSubsections () const |
| Non-mutable access to the registered subsections. | |
 Public Member Functions inherited from ProgressLogger Public Member Functions inherited from ProgressLogger | |
| ProgressLogger () | |
| Constructor. | |
| virtual | ~ProgressLogger () |
| Destructor. | |
| ProgressLogger (const ProgressLogger &other) | |
| Copy constructor. | |
| ProgressLogger & | operator= (const ProgressLogger &other) |
| Assignment Operator. | |
| void | setLogType (LogType type) const |
| Sets the progress log that should be used. The default type is NONE! | |
| LogType | getLogType () const |
| Returns the type of progress log being used. | |
| void | setLogger (ProgressLoggerImpl *logger) |
| Sets the logger to be used for progress logging. | |
| void | startProgress (SignedSize begin, SignedSize end, const String &label) const |
| Initializes the progress display. | |
| void | setProgress (SignedSize value) const |
| Sets the current progress. | |
| void | endProgress (UInt64 bytes_processed=0) const |
| void | nextProgress () const |
| increment progress by 1 (according to range begin-end) | |
Protected Member Functions | |
| std::vector< MultiplexIsotopicPeakPattern > | generatePeakPatterns_ (int charge_min, int charge_max, int peaks_per_peptide_max, const std::vector< MultiplexDeltaMasses > &mass_pattern_list) |
| generate list of m/z shifts | |
| void | correctPeptideIntensities_ (const MultiplexIsotopicPeakPattern &pattern, std::map< size_t, SplinePackage > &spline_chromatograms, const std::vector< double > &rt_peptide, std::vector< double > &intensity_peptide) const |
| determine ratios through linear regression and correct peptide intensities | |
| std::vector< double > | determinePeptideIntensitiesCentroided_ (const MultiplexIsotopicPeakPattern &pattern, const std::multimap< size_t, MultiplexSatelliteCentroided > &satellites) |
| calculate peptide intensities | |
| std::vector< double > | determinePeptideIntensitiesProfile_ (const MultiplexIsotopicPeakPattern &pattern, const std::multimap< size_t, MultiplexSatelliteProfile > &satellites) |
| calculate peptide intensities | |
| void | generateMapsCentroided_ (const std::vector< MultiplexIsotopicPeakPattern > &patterns, const std::vector< MultiplexFilteredMSExperiment > &filter_results, std::vector< std::map< int, GridBasedCluster > > &cluster_results) |
| generates consensus and feature maps containing all peptide multiplets | |
| void | generateMapsProfile_ (const std::vector< MultiplexIsotopicPeakPattern > &patterns, const std::vector< MultiplexFilteredMSExperiment > &filter_results, const std::vector< std::map< int, GridBasedCluster > > &cluster_results) |
| generates consensus and feature maps containing all peptide multiplets | |
 Protected Member Functions inherited from DefaultParamHandler Protected Member Functions inherited from DefaultParamHandler | |
| virtual void | updateMembers_ () |
| This method is used to update extra member variables at the end of the setParameters() method. | |
| void | defaultsToParam_ () |
| Updates the parameters after the defaults have been set in the constructor. | |
Protected Attributes | |
| MSExperiment | exp_profile_ |
| MSExperiment | exp_centroid_ |
| bool | centroided_ |
| ProgressLogger | prog_log_ |
| bool | progress_ |
| unsigned | charge_min_ |
| unsigned | charge_max_ |
| unsigned | isotopes_per_peptide_min_ |
| unsigned | isotopes_per_peptide_max_ |
| std::map< String, double > | label_mass_shift_ |
| FeatureMap | feature_map_ |
| ConsensusMap | consensus_map_ |
| MSExperiment | exp_blacklist_ |
 Protected Attributes inherited from DefaultParamHandler Protected Attributes inherited from DefaultParamHandler | |
| Param | param_ |
| Container for current parameters. | |
| Param | defaults_ |
| Container for default parameters. This member should be filled in the constructor of derived classes! | |
| std::vector< String > | subsections_ |
| Container for registered subsections. This member should be filled in the constructor of derived classes! | |
| String | error_name_ |
| Name that is displayed in error messages during the parameter checking. | |
| bool | check_defaults_ |
| If this member is set to false no checking if parameters in done;. | |
| bool | warn_empty_defaults_ |
| If this member is set to false no warning is emitted when defaults are empty;. | |
 Protected Attributes inherited from ProgressLogger Protected Attributes inherited from ProgressLogger | |
| LogType | type_ |
| time_t | last_invoke_ |
| ProgressLoggerImpl * | current_logger_ |
Additional Inherited Members | |
 Public Types inherited from ProgressLogger Public Types inherited from ProgressLogger | |
| enum | LogType { CMD , GUI , NONE } |
| Possible log types. More... | |
 Static Public Member Functions inherited from DefaultParamHandler Static Public Member Functions inherited from DefaultParamHandler | |
| static void | writeParametersToMetaValues (const Param &write_this, MetaInfoInterface &write_here, const String &key_prefix="") |
| Writes all parameters to meta values. | |
 Static Protected Attributes inherited from ProgressLogger Static Protected Attributes inherited from ProgressLogger | |
| static int | recursion_depth_ |
FeatureFinderMultiplexAlgorithm is a tool for the fully automated analysis of quantitative proteomics data. It detects pairs of isotopic envelopes with fixed m/z separation. It requires no prior sequence identification of the peptides and works on both profile or centroided spectra. In what follows we outline the algorithm.
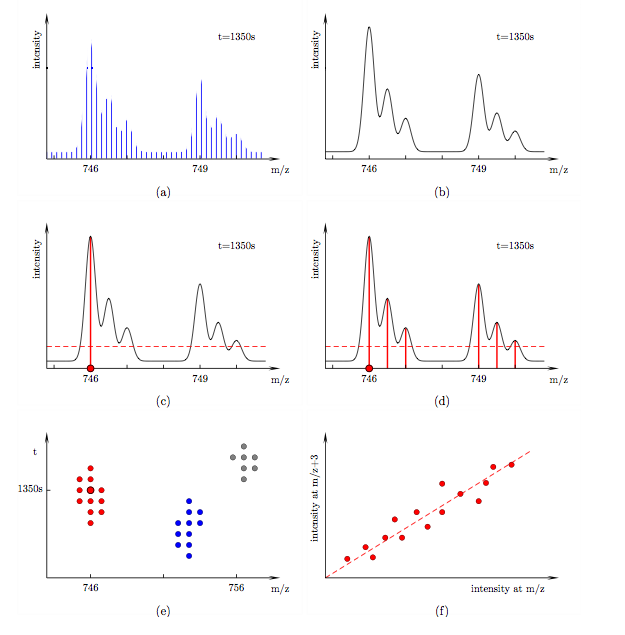
Algorithm The algorithm is divided into three parts: filtering, clustering and linear fitting, see Fig. (d), (e) and (f). In the following discussion let us consider a particular mass spectrum at retention time 1350 s, see Fig. (a). It contains a peptide of mass 1492 Da and its 6 Da heavier labelled counterpart. Both are doubly charged in this instance. Their isotopic envelopes therefore appear at 746 and 749 in the spectrum. The isotopic peaks within each envelope are separated by 0.5. The spectrum was recorded at finite intervals. In order to read accurate intensities at arbitrary m/z we spline-fit over the data, see Fig. (b). We would like to search for such peptide pairs in our LC-MS data set. As a warm-up let us consider a standard intensity cut-off filter, see Fig. (c).
Scanning through the entire m/z range (red dot) only data points with intensities above a certain threshold pass the filter. Unlike such a local filter, the filter used in our algorithm takes intensities at a range of m/z positions into account, see Fig. (d). A data point (red dot) passes if
Let us now filter not only a single spectrum but all spectra in our data set. Data points that pass the filter form clusters in the t-m/z plane, see Fig. (e). Each cluster corresponds to the mono-isotopic mass trace of the lightest peptide of a SILAC pattern. We now use hierarchical clustering methods to assign each data point to a specific cluster. The optimum number of clusters is determined by maximizing the silhouette width of the partitioning. Each data point in a cluster corresponds to three pairs of intensities (at [m/z, m/z+3], [m/z+0.5, m/z+3.5] and [m/z+1, m/z+4]). A plot of all intensity pairs in a cluster shows a clear linear correlation, see Fig. (f). Using linear regression we can determine the relative amounts of labelled and unlabelled peptides in the sample.
default constructor
|
protected |
determine ratios through linear regression and correct peptide intensities
In most labelled mass spectrometry experiments, the fold change i.e. ratio and not the individual peptide intensities are of primary interest. For that reason, we determine the ratios from interpolated chromatogram data points directly, and then correct the current ones.
| [in] | pattern | Isotopic peak pattern |
| [in,out] | spline_chromatograms | Spline chromatograms to be used/modified |
| [in] | rt_peptide | Retention times of peptides |
| [out] | intensity_peptide | Corrected peptide intensities |
|
protected |
calculate peptide intensities
| [in] | pattern | Isotopic peak pattern |
| [in] | satellites | Satellite peaks |
|
protected |
calculate peptide intensities
| [in] | pattern | Isotopic peak pattern |
| [in] | satellites | Satellite peaks |
|
protected |
generates consensus and feature maps containing all peptide multiplets
| [in] | patterns | patterns of isotopic peaks we have been searching for |
| [in] | filter_results | filter results for each of the patterns |
| [in,out] | cluster_results | clusters of filter results |
|
protected |
generates consensus and feature maps containing all peptide multiplets
| [in] | patterns | patterns of isotopic peaks we have been searching for |
| [in] | filter_results | filter results for each of the patterns |
| [in] | cluster_results | clusters of filter results |
|
protected |
generate list of m/z shifts
| [in] | charge_min | minimum charge |
| [in] | charge_max | maximum charge |
| [in] | peaks_per_peptide_max | maximum number of isotopes in peptide |
| [in] | mass_pattern_list | mass shifts due to labelling |
| MSExperiment & getBlacklist | ( | ) |
| ConsensusMap & getConsensusMap | ( | ) |
| FeatureMap & getFeatureMap | ( | ) |
get methods
| void run | ( | MSExperiment & | exp, |
| bool | progress | ||
| ) |
main method for feature detection
|
protected |
|
protected |
|
protected |
|
protected |
|
protected |
|
protected |
|
protected |
|
protected |
|
protected |
|
protected |
|
protected |
|
protected |
|
protected |