#include <BALL/QSAR/Model.h>
Definition at line 34 of file Model.h.
| BALL::QSAR::Model::Model |
( |
const QSARData & |
q | ) |
|
constructur
- Parameters
-
| q | QSARData object, from which the data for this model should be taken |
| virtual BALL::QSAR::Model::~Model |
( |
| ) |
|
|
virtual |
| void BALL::QSAR::Model::addLambda |
( |
Eigen::MatrixXd & |
matrix, |
|
|
double & |
lambda |
|
) |
| |
|
protected |
adds offset lambda to the diagonal of the given matrix
| void BALL::QSAR::Model::backTransformPrediction |
( |
Eigen::VectorXd & |
pred | ) |
|
|
protected |
transforms a prediction (obtained by Model.train()) according to the inverse of the transformation(s) of the activity values of the training data
| virtual double BALL::QSAR::Model::calculateStdErr |
( |
| ) |
|
|
inlinevirtual |
| void BALL::QSAR::Model::copyData |
( |
const Model & |
m | ) |
|
copies the data (descriptor matrix, names of substances and descriptors) and the IDs of the selected descriptors from m
| void BALL::QSAR::Model::copyDescriptorIDs |
( |
const Model & |
m | ) |
|
copies the IDs of the selected descriptors from m
| void BALL::QSAR::Model::deleteDescriptorIDs |
( |
| ) |
|
removes all entries from descriptor_IDs
| std::multiset<unsigned int>* BALL::QSAR::Model::getDescriptorIDs |
( |
| ) |
|
returns a const pointer to the descriptor IDs of this model
| const Eigen::MatrixXd* BALL::QSAR::Model::getDescriptorMatrix |
( |
| ) |
|
returns a const pointer to the descriptor matrix of this model
| const vector<string>* BALL::QSAR::Model::getDescriptorNames |
( |
| ) |
|
returns a const pointer to the names of the descriptors of this model
| const Eigen::MatrixXd BALL::QSAR::Model::getDescriptorTransformations |
( |
| ) |
|
returns descriptor transformations
| virtual vector<double> BALL::QSAR::Model::getParameters |
( |
| ) |
const |
|
virtual |
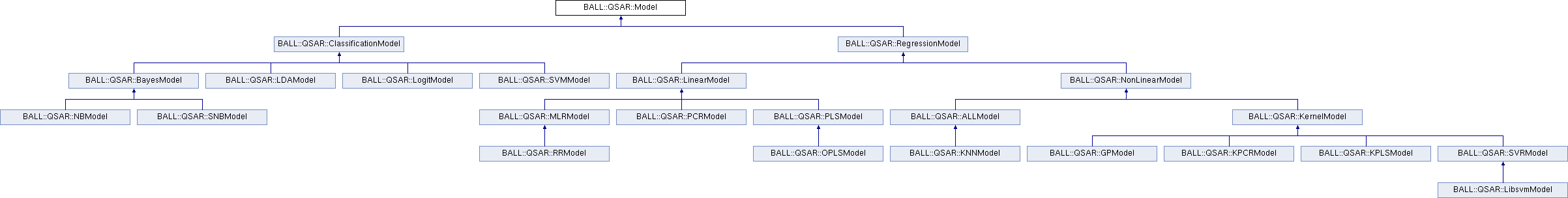
Reimplemented in BALL::QSAR::KPLSModel, BALL::QSAR::PLSModel, BALL::QSAR::KPCRModel, BALL::QSAR::GPModel, BALL::QSAR::PCRModel, BALL::QSAR::NBModel, BALL::QSAR::OPLSModel, BALL::QSAR::ALLModel, BALL::QSAR::SNBModel, BALL::QSAR::RRModel, BALL::QSAR::LDAModel, BALL::QSAR::LibsvmModel, and BALL::QSAR::KNNModel.
| const vector<string>* BALL::QSAR::Model::getSubstanceNames |
( |
| ) |
|
returns a const pointer to the names of the substances of this model
| Eigen::VectorXd BALL::QSAR::Model::getSubstanceVector |
( |
const vector< double > & |
substance, |
|
|
bool |
transform |
|
) |
| |
|
protected |
returns a Row-Vector containing only the values for these descriptors, that have been selected for this model
- Parameters
-
| substance | a vector of all descriptor values for the substance to be predicted |
| Eigen::VectorXd BALL::QSAR::Model::getSubstanceVector |
( |
const Eigen::VectorXd & |
substance, |
|
|
bool |
transform |
|
) |
| |
|
protected |
| const string* BALL::QSAR::Model::getType |
( |
| ) |
|
returns the type of the current model, e.g. "MLR", "PLS", ...
| void BALL::QSAR::Model::getUnnormalizedFeatureValue |
( |
int |
compound, |
|
|
int |
feature, |
|
|
double & |
return_value |
|
) |
| |
Fetches the un-normalized value for the specified feature of the desired compound (instance) from the data that this Model currently contains. This method is needed for visualization purposes only.
| void BALL::QSAR::Model::getUnnormalizedResponseValue |
( |
int |
compound, |
|
|
int |
response, |
|
|
double & |
return_value |
|
) |
| |
Fetches the un-normalized value for the specified response of the desired compound (instance) from the data that this Model currently contains. This method is needed for visualization purposes only.
| const Eigen::MatrixXd* BALL::QSAR::Model::getY |
( |
| ) |
|
returns a const pointer to the activity values of this model
| const Eigen::MatrixXd BALL::QSAR::Model::getYTransformations |
( |
| ) |
|
| virtual void BALL::QSAR::Model::operator= |
( |
const Model & |
m | ) |
|
|
virtual |
copy constructur; creates a model with the same specifications as the given one (same model and kernel parameters). If the given model has been trained, the training result is copied as well.
Note, that the input data that has been read by m to m.descriptor_matrix_ and m.Y_ is NOT copied to new model, since the input data is not part of the specification of a model. If nevertheless, copying of the input data is desired, use function copyData() (afterwards).
Reimplemented in BALL::QSAR::KernelModel.
| virtual bool BALL::QSAR::Model::optimizeParameters |
( |
int |
, |
|
|
int |
|
|
) |
| |
|
inlinevirtual |
optimizes parameters (!=number of features) of the model, e.g. no of latente variables in case of PLS model or kernel width in case of automated lazy learning model.
The number of selected features (=descriptors) is NOT changed by this method. Use class FeatureSelection in order to do this.
- Returns
- 1 if parameters were optimized using cross-validation. The best Q2 value is assumed to be saved in ModelValidation.Q2
0 if the model has no parameters to be optimized, so that no cross-validation was done.
Reimplemented in BALL::QSAR::KPLSModel, BALL::QSAR::PLSModel, BALL::QSAR::OPLSModel, BALL::QSAR::ALLModel, and BALL::QSAR::KNNModel.
Definition at line 89 of file Model.h.
| bool BALL::QSAR::Model::optimizeParameters |
( |
int |
k | ) |
|
| virtual Eigen::VectorXd BALL::QSAR::Model::predict |
( |
const vector< double > & |
substance, |
|
|
bool |
transform |
|
) |
| |
|
pure virtual |
Predicts the activities of a given substance
- Parameters
-
| substance | the substance which activity is to be predicted in form of a vecor containing the values for all descriptors (if neccessary, relevant descriptors will be selected automatically) |
| transform | determines whether the values for each descriptor of the given substance should be transformed before prediction of activity.
If (transform==1): each descriptor value is transformed according to the centering of the respective column of QSARData.descriptor_matrix used to train this model.
If the substance to be predicted is part of the same input data (e.g. same SD-file) as the training data (as is the case during cross validation), transform should therefore be set to 0. |
- Returns
- a RowVector containing one value for each predicted activity
Implemented in BALL::QSAR::KernelModel, BALL::QSAR::GPModel, BALL::QSAR::NBModel, BALL::QSAR::SNBModel, BALL::QSAR::LDAModel, BALL::QSAR::LinearModel, BALL::QSAR::LogitModel, and BALL::QSAR::ALLModel.
| void BALL::QSAR::Model::readDescriptorInformation |
( |
| ) |
|
|
protected |
reads selected descriptors, their names and the information about their transformations (mean and stddev of each descriptor). This function is used after feature selection to read information about the selected features
| virtual void BALL::QSAR::Model::readDescriptorInformationFromFile |
( |
std::ifstream & |
in, |
|
|
int |
no_descriptors, |
|
|
bool |
transformation |
|
) |
| |
|
protectedvirtual |
| virtual void BALL::QSAR::Model::readFromFile |
( |
string |
filename | ) |
|
|
pure virtual |
| void BALL::QSAR::Model::readMatrix |
( |
Eigen::MatrixXd & |
mat, |
|
|
std::ifstream & |
in, |
|
|
unsigned int |
lines, |
|
|
unsigned int |
col |
|
) |
| |
|
protected |
reconstructs a Eigen::MatrixXd from a given input stream after resizing the given Eigen::MatrixXd as specified
| void BALL::QSAR::Model::readModelParametersFromFile |
( |
std::ifstream & |
in | ) |
|
|
protected |
| void BALL::QSAR::Model::readResponseTransformationFromFile |
( |
std::ifstream & |
in, |
|
|
int |
no_y |
|
) |
| |
|
protected |
| void BALL::QSAR::Model::readTrainingData |
( |
| ) |
|
copies the data for the relevant descriptors from the bound QSARData object into this model and updates Model.descriptor_transformations and Model.y_transformations .
If no explicit feature selection was done, i.e. if descriptor_IDs is emtpy, all data is fetched.
If feature selection was done, i.e. if descriptor_IDs is not empty, only the columns of the relevant descriptors are fetched.
| void BALL::QSAR::Model::readVector |
( |
Eigen::RowVectorXd & |
vec, |
|
|
std::ifstream & |
in, |
|
|
unsigned int |
no_cells, |
|
|
bool |
column_vector |
|
) |
| |
|
protected |
| virtual void BALL::QSAR::Model::saveDescriptorInformationToFile |
( |
std::ofstream & |
out | ) |
|
|
protectedvirtual |
| void BALL::QSAR::Model::saveModelParametersToFile |
( |
std::ofstream & |
out | ) |
|
|
protected |
| void BALL::QSAR::Model::saveResponseTransformationToFile |
( |
std::ofstream & |
out | ) |
|
|
protected |
| virtual void BALL::QSAR::Model::saveToFile |
( |
string |
filename | ) |
|
|
pure virtual |
| void BALL::QSAR::Model::setDataSource |
( |
const QSARData * |
q | ) |
|
| void BALL::QSAR::Model::setDescriptorIDs |
( |
const std::multiset< unsigned int > & |
sl | ) |
|
manually specify a set of descriptors
| virtual void BALL::QSAR::Model::setParameters |
( |
vector< double > & |
| ) |
|
|
inlinevirtual |
sets the model parameters according to the given values.
Reimplemented in BALL::QSAR::KPLSModel, BALL::QSAR::PLSModel, BALL::QSAR::NBModel, BALL::QSAR::KPCRModel, BALL::QSAR::GPModel, BALL::QSAR::PCRModel, BALL::QSAR::OPLSModel, BALL::QSAR::ALLModel, BALL::QSAR::SNBModel, BALL::QSAR::RRModel, BALL::QSAR::LDAModel, BALL::QSAR::LibsvmModel, and BALL::QSAR::KNNModel.
Definition at line 97 of file Model.h.
| virtual void BALL::QSAR::Model::train |
( |
| ) |
|
|
pure virtual |
Starts training the model.
Implemented in BALL::QSAR::KPCRModel, BALL::QSAR::KPLSModel, BALL::QSAR::PLSModel, BALL::QSAR::PCRModel, BALL::QSAR::GPModel, BALL::QSAR::NBModel, BALL::QSAR::MLRModel, BALL::QSAR::SNBModel, BALL::QSAR::RRModel, BALL::QSAR::ALLModel, BALL::QSAR::OPLSModel, BALL::QSAR::LDAModel, BALL::QSAR::LibsvmModel, and BALL::QSAR::LogitModel.
pointer to the input data class for this model
Definition at line 147 of file Model.h.
| int BALL::QSAR::Model::default_no_opt_steps_ |
|
protected |
The default number of steps for model parameter optimization.
It can be adjusted by the different types of models.
Standard default value is 30.
Definition at line 159 of file Model.h.
| std::multiset<unsigned int> BALL::QSAR::Model::descriptor_IDs_ |
|
protected |
list containing the IDs of the selected descriptors (=features); with IDs >= 0
If this list is empty, it is assumed that no feature selection was done, i.e. that all descriptors are to be considered for cross-validation and prediction of activity.
If it is not empty, only the descriptors in this list are used for cross-validation and prediction of activity.
Definition at line 232 of file Model.h.
| Eigen::MatrixXd BALL::QSAR::Model::descriptor_matrix_ |
|
protected |
matrix containing the values of each descriptor for each substance
Definition at line 206 of file Model.h.
| vector<string> BALL::QSAR::Model::descriptor_names_ |
|
protected |
names of all descriptors
Definition at line 212 of file Model.h.
| Eigen::MatrixXd BALL::QSAR::Model::descriptor_transformations_ |
|
protected |
2xm dimensional matrix (m=no of descriptors) containing mean and stddev of each selected descriptor.
The content of this matrix is updated only by Model.readTrainingData()
Definition at line 216 of file Model.h.
a ModelValidation object, that is used to validate this model and that will contain the results of the validations
Definition at line 150 of file Model.h.
| vector<string> BALL::QSAR::Model::substance_names_ |
|
protected |
names of all substances
Definition at line 209 of file Model.h.
| String BALL::QSAR::Model::type_ |
|
protected |
The type of model, e.g. "MLR", "GP", ...
Definition at line 227 of file Model.h.
| Eigen::MatrixXd BALL::QSAR::Model::Y_ |
|
protected |
Matrix containing the experimentally determined results (active/non-active) for each substance.
Each column contains the values for one activity.
Definition at line 224 of file Model.h.
| Eigen::MatrixXd BALL::QSAR::Model::y_transformations_ |
|
protected |
2xc dimensional matrix (c=no of activities) containing mean and stddev of each activity.
The content of this matrix is updated only by Model.readTrainingData()
Definition at line 220 of file Model.h.
 1.8.7
1.8.7